Function for printing the summary of species-area relationship objects from
the SSARP::create_SAR() function
Usage
# S3 method for class 'SAR'
print(x, printlen = NULL, ...)Examples
# The GBIF key for the Anolis genus is 8782549
# Obtained with: key <- get_key(query = "Anolis", rank = "genus")
key <- 8782549
# Read in example dataset obtained through:
# dat <- get_data(key = key, limit = 100)
dat <- read.csv(system.file("extdata",
"SSARP_Example_Dat.csv",
package = "SSARP"))
occs <- find_land(occurrences = dat)
areas <- find_areas(occs = occs)
#> ℹ Recording island names...
#> ℹ Assembling island dictionary...
#> ℹ Adding areas to final dataframe...
seg <- create_SAR(areas, npsi = 0)
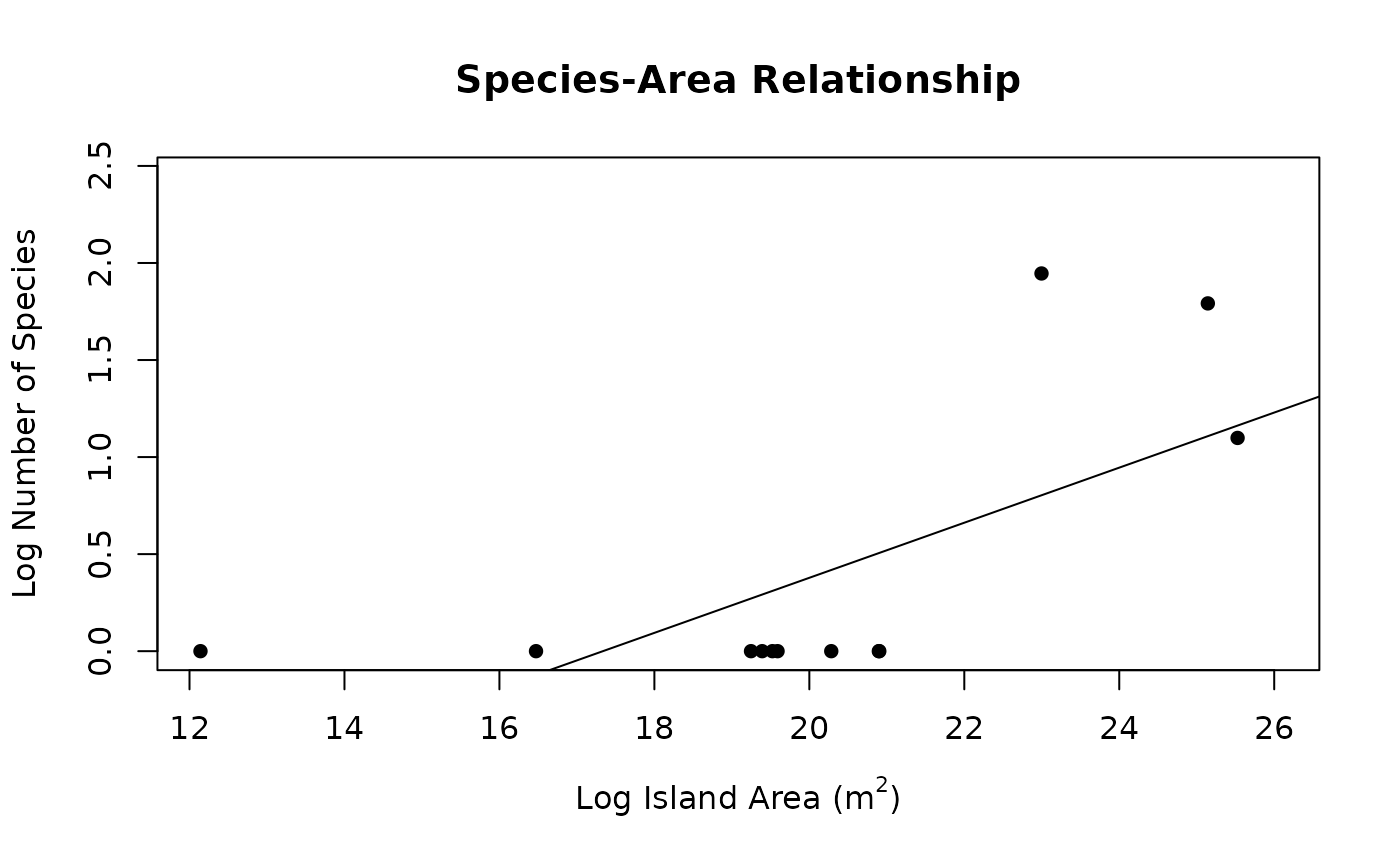 print(seg)
#>
#> Call:
#> stats::lm(formula = y ~ x, data = dat)
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -0.5061 -0.3444 -0.2814 0.2629 1.1428
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) -2.45955 0.99560 -2.470 0.0331 *
#> x 0.14188 0.04864 2.917 0.0154 *
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Residual standard error: 0.5813 on 10 degrees of freedom
#> Multiple R-squared: 0.4597, Adjusted R-squared: 0.4057
#> F-statistic: 8.509 on 1 and 10 DF, p-value: 0.01538
#>
print(seg)
#>
#> Call:
#> stats::lm(formula = y ~ x, data = dat)
#>
#> Residuals:
#> Min 1Q Median 3Q Max
#> -0.5061 -0.3444 -0.2814 0.2629 1.1428
#>
#> Coefficients:
#> Estimate Std. Error t value Pr(>|t|)
#> (Intercept) -2.45955 0.99560 -2.470 0.0331 *
#> x 0.14188 0.04864 2.917 0.0154 *
#> ---
#> Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
#>
#> Residual standard error: 0.5813 on 10 degrees of freedom
#> Multiple R-squared: 0.4597, Adjusted R-squared: 0.4057
#> F-statistic: 8.509 on 1 and 10 DF, p-value: 0.01538
#>