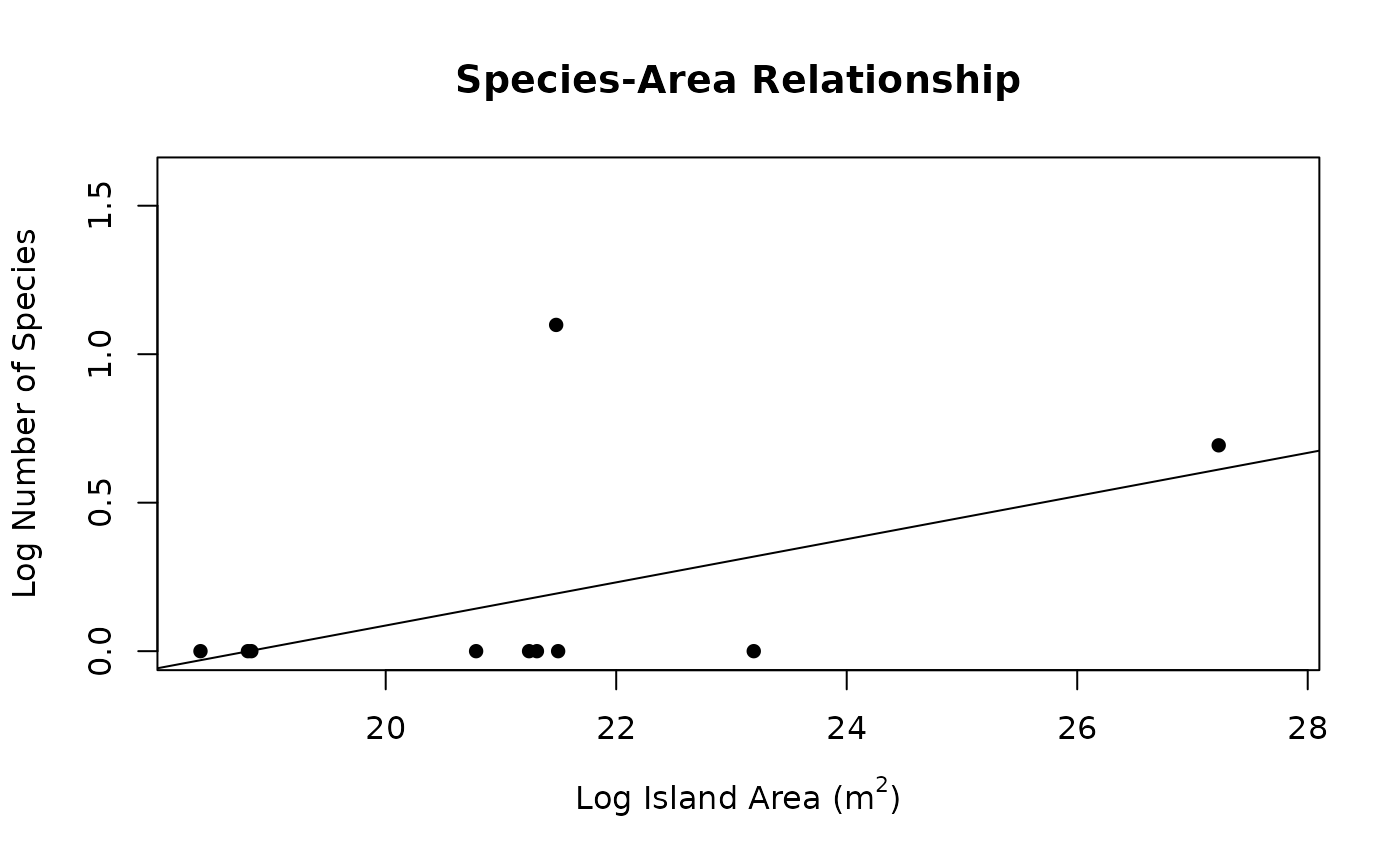
Use the SSARP workflow for creating a species-area relationship for island species using occurrence data from GBIF without having to go through every individual step.
Arguments
- taxon
The name of the taxon of interest.
- rank
The taxonomic rank associated with the taxon of interest.
- limit
The number of GBIF occurrence records to obtain.
- geometry
(character) A polygon written in Well Known Text (WKT) format to pass to the
SSARP::get_data()function. If no polygon is given, records from across the globe will be returned.- continent
(logical) If TRUE, continental areas are removed from the species-area relationship data. If FALSE, continental areas are not removed from the species-area relationship data. Default: TRUE
- npsi
The number of breakpoints to estimate. Default: 1
Value
A list of 3 including: the summary output, the segmented regression object, and the aggregated dataframe used to create the plot.
Examples
seg <- quick_create_SAR(taxon = "Phelsuma", rank = "genus", limit = 100, npsi = 0)
#> ℹ Finding taxon key
#> ℹ Gathering data
#> ℹ Finding land
#> ℹ Gathering areas
#> ℹ Recording island names...
#> ℹ Assembling island dictionary...
#> ℹ Adding areas to final dataframe...
#> ℹ Removing continental areas